| Editorial Commentary | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Microbiological study of diabetic foot infections | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sivaraman Umadevi, Shailesh Kumar, Noyal Mariya Joseph, Joshy M Easow, G Kandhakumari, Sreenivasan Srirangaraj,
Sruthi Raj, Selvaraj Stephen
* Department of Microbiology, Mahatma Gandhi Medical College and Research Institute, Pondicherry, India.
Corresponding Author: Dr. Uma Devi S., Department of Microbiology, Mahatma Gandhi Medical College and Research Institute, Pillaiyarkuppam, Pondicherry – 607 402. India. Email: drumadevi@yahoo.co.in
Diabetic foot is one of the most feared complications of diabetes and is the leading cause of hospitalisation in diabetic patients. Diabetic foot is characterised by several pathological complications such as neuropathy, peripheral vascular disease, foot ulceration and infection with or without osteomyelitis, leading to development of gangrene and even necessitating limb amputation [1,2]. Diabetic patients have a lifetime risk as high as 25% for developing foot ulceration [3]. Diabetic ulcers have 15 to 46 times higher risk of limb amputation than foot ulcers due to other causes [4]. Every year more than a million diabetic patients require limb amputation [1]. The impaired
micro-vascular circulation in patients with diabetic foot limits the
access of phagocytes favouring development of infection [2,5]. Escherichia coli, Proteus spp., Pseudomonas spp., Staphylococcus aureus and Enterococcus spp. are
the most frequent pathogens contributing to
progressive and widespread tissue destruction
[2,5]. Diabetic foot infections are often
polymicrobial [4,5]. Methicillin-resistant Staphylococcus aureus (MRSA) has been commonly
isolated from 10-40% of the diabetic wounds [6,7,8].
The increasing association of multi-drug resistant
(MDR) pathogens with diabetic foot ulcers further
compounds the challenge faced by the physician or
the surgeon in treating diabetic ulcers without
resorting to amputation [9]. Infection with MDR
pathogens is also responsible for the increased
duration of hospitalisation, cost of management, morbidity and mortality of the diabetic patients [5]. So, this study was performed to determine the common etiological agents of diabetic foot infections in a tertiary hospital and their in vitro susceptibility to routinely used antibiotics. The prevalence of MDR pathogens in patients with diabetic foot infections was also studied. Methods A prospective study was performed over a period of one year from September 2008 to August 2009. The study was conducted at a tertiary care teaching hospital in Pondicherry, India. All patients with diabetic foot infections were included in the study. Processing of specimens- Pus or discharges from the ulcer base and debrided necrotic tissue were obtained. The specimens were taken immediately to the microbiology laboratory and processed without any delay. The specimens were subjected to Gram staining and were simultaneously inoculated on blood agar and Mac Conkey agar for isolation of o aerobic bacteria. After 24 hours incubation at 37 C, the bacterial isolates were identified based on standard bacteriological methods [10]. Antibiotic susceptibility testing- Antibiotic susceptibility testing was performed by Kirby Bauer’s disc diffusion method according to Clinical Laboratory Standards Institute (CLSI) guidelines [11]. Amoxicillin-clavulanic acid, piperacillintazobactam, tetracycline, ciprofloxacin, trimethoprim-sulfamethoxazole, gentamicin, amikacin, cefuroxime, ceftriaxone and imipenem were tested for Enterobacteriaceae. Piperacillintazobactam, ciprofloxacin, gentamicin, amikacin, netilmicin, ceftazidime and imipenem were tested for Pseudomonas species. Piperacillin-tazobactam, tetracycline, ciprofloxacin, trimethoprimsulfamethoxazole, gentamicin, amikacin, ceftriaxone, ceftazidime and imipenem were tested for Acinetobacter species. Penicillin, amoxicillinclavulanic acid, erythromycin, trimethoprim-sulfamethoxazole, tetracycline, ciprofloxacin, gentamicin, ceftriaxone and vancomycin were tested for Staphylococcus species. Penicillin, erythromycin, tetracycline, ciprofloxacin, high level gentamicin and vancomycin were tested for Enterococcus species. MRSA, vancomycin-resistant enterococci (VRE), Gram-negative bacilli producing ESBL, MDR P. aeruginosa (resistant to ≥ 3 anti-pseudomonal classes of antimicrobial agents) and MDR Acinetobacter spp. (resistant to classes of antimicrobial agents) are defined as multi-drug resistant (MDR) pathogens [12,13,14]. Combination disc method using both cefotaxime and ceftazidime, alone and in combination with clavulanic acid was performed for detection of extended spectrum. β-lactamase (ESBL) among the members of Enterobacteriaceae [15]. Five mm or more increase in zone of inhibition for either cefotaxime-clavulanic acid or ceftazidimeclavulanic acid disc compared to the cefotaxime or ceftazidime disc respectively was taken as confirmatory evidence of ESBL production. Staphylococcus aureus isolates were screened for methicillin resistance using oxacillin-salt screen agar containing 6μg/mL oxacillin and 4% NaCl according to CLSI guidelines [11]. Results Of the 105 patients with diabetic foot, 84 (80%) were male and 21 (20%) were female. The age ranged from 32 to 73 years with mean age being 47 ± 11 years. A total of 171 bacteria were isolated from these 105 patients. The bacteria isolated from the diabetic foot ulcers are summarised in Table 1. In 47 (44.8%) patients only one pathogen was isolated, while in 55 (52.4%) patients more than one pathogen was isolated (41 were infected with two pathogens, while 14 had three pathogens). In 3 (2.9%) patients, no isolate was obtained. Gram-positive organisms were found as the only isolate in 9 (8.6%) patients, while 55 (52.4%) patients had only gram-negative organisms. The remaining 41 patients (39.0%) had both gram-positive and gram-negative organisms. The ratio of gram-negative to gram-positive organisms isolated from diabetic foot ulcers was 2.4:1. Gram-negative bacteria accounted for 70.8%, while gram-positive bacteria accounted for 29.2%. Table 1- Bacteria isolated from diabetic foot ulcers
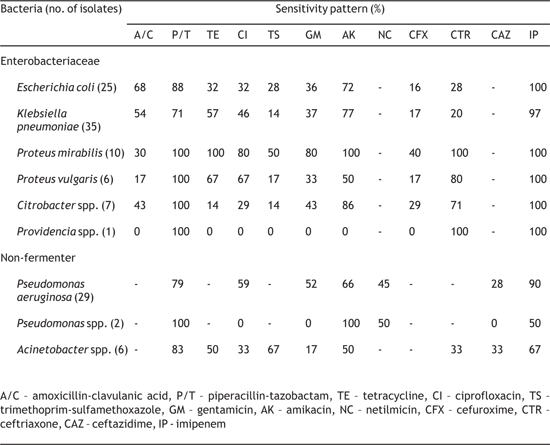
The sensitivity of the isolated gram-negative bacteria to commonly used antibiotics is summarised in Table 2. Majority of isolates of Escherichia coli and Klebsiella pneumoniae were susceptible to amikacin, piperacillin-tazobactam and imipenem, but resistant to other antibiotics tested except amoxicillin-clavulanic acid for which they were showing variable susceptibility. Similarly, most of our Proteus spp. were susceptible to tetracycline, ciprofloxacin, amikacin, ceftriaxone, piperacillin-tazobactam and imipenem, while being less susceptible to amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole and cefuroxime. However, Proteus mirabilis was relatively more susceptible than Proteus vulgaris to most antibiotics. Citrobacter spp. were susceptible to piperacillin-tazobactam, amikacin, ceftriaxone and imipenem, but resistant to other antibiotics tested. Table 2- Sensitivity pattern of Gram negative bacteria isolated from diabetic foot ulcer
Most of the Pseudomonas aeruginosa were susceptible to piperacillin-tazobactam and imipenem, while they were showing varying susceptibility to ciprofloxacin, gentamicin, amikacin and netilmicin. Similarly, majority of Acinetobacter spp. were susceptible to piperacillintazobactam, imipenem and trimethoprimsulfamethoxazole, while being less susceptible to gentamicin, amikacin, ciprofloxacin, tetracycline, ceftiaxone and ceftazidime. The antibiotic susceptibility patterns of the grampositive
bacteria isolated from diabetic ulcers are
shown in Table 3. Staphylococcus aureus were most
often susceptible to erythromycin, tetracycline and
vancomycin, but were relatively less susceptible to
amoxicillin-clavulanic acid, trimethoprimsulfamethoxazole,
ciprofloxacin, gentamicin and ceftriaxone. None of the Staphylococcus aureus were susceptible to penicillin. Most of the Enterococcus spp. were susceptible only to
vancomycin. However they showed varying
susceptibility to tetracycline, penicillin, and
ciprofloxacin. High-level aminoglycoside resistance was observed in 33% of the Enterococcus spp. Table 3- Sensitivity pattern of Gram positive bacteria isolated from diabetic foot ulcer
Nineteen of the 29 (65.5%) Staphylococcus aureus were resistant to oxacillin and were therefore considered as methicillin resistant Staphylococcus aureus (MRSA). ESBL production was detected in 47 of the 84 (56%) isolates belonging to Enterobacteriaceae. Proteus spp. (10 out of 16 isolates, 62.5%), Klebsiella pneumoniae (21 out of 35 isolates, 60%) and Escherichia coli (14 out of 25 isolates, 56%) were frequently ESBL producers. Twenty two multi-drug resistant non-fermenting gram-negative bacteria such as Pseudomonas spp. and Acinetobacter spp. were observed in our study. Overall 89 of the 171 (52%) isolates were MDR pathogens. Discussion Diabetic patients often have chronic non-healing foot ulcers due to several underlying factors such as neuropathy, high plantar pressures and peripheral arterial disease [16]. Such chronic long-standing ulcers are more prone for infection which further delays the wound healing process. A wide range of bacteria can cause infection in these patients. In this study, gram-negative bacteria were the predominant pathogens, Klebsiella pneumoniae being the commonest aetiological agent, followed by Pseudomonas aeruginosa and Staphylococcusaureus. Similarly, in two recent studies, gramnegative bacteria were the commonest agents [5,7]. But earlier studies have documented grampositive bacteria as the predominant organisms associated with diabetic foot infections [17,18]. Therefore, there seems to be a changing trend in the organisms causing diabetic foot infections, with gram-negative bacteria replacing gram-positive bacteria as commonest agents. Polymicrobial infection was observed in 52% patients, which is similar to other studies [2,4,7]. The awareness about the antibiotic susceptibility pattern of the isolates from diabetic foot infections is crucial for appropriate treatment of cases. Although in an earlier Indian study, all members of Enterobacteriaceae were found to be uniformly sensitive to gentamicin and ciprofloxacin [2], in the present study most of them except Proteus spp. were resistant to these antibiotics. Another recent study also has reported increasing resistance to these drugs [5]. Therefore, empirical use of these antibiotics in diabetic foot infections should not be advocated. However, members of Enterobacteriaceae were found to be susceptible to amikacin, piperacillin-tazobactam and imipenem. Similarly, inarecentIndianstudy, Enterobacteriaceae were found to be sensitive to ticarcillin-clavulanic acid, cefoperazone-sulbactam and imipenem [5]. So, empirical treatment of diabetic foot infections in areas with increased drug resistance should include a combination of these antibiotics. Increased resistance to cefuroxime and ceftriaxone was noted among the Klebsiella pneumoniae andEscherichia coli isolated in the study group. Although production of ESBL can explain the resistance in many of the isolates, but some of the resistant isolates did not produce ESBL. Production of other enzymes such as AmpC β-lactamases, capable of hydrolysing the extended-spectrum cephalosporins (cefuroxime, cefotaxime, ceftriaxone, ceftazidime) could be the reason for resistance in non-ESBL producing isolates [19]. In the present study, Proteus spp. were susceptibile to ceftriaxone but were often resistant to cefuroxime, a second generation cephalosporin. This could be explained by the fact that Proteus spp. are known to produce unique β-lactamase (cefuroximase) that has high activity mainly against cefuroxime and cefotaxime [20]. It was observed that 56% of the members of Enterobacteriaceae were producing ESBL. Similarly, Gadepalli et al also have documented ESBL production in 44.7% of bacterial isolates [5]. ESBL producers are resistant to all extended-spectrum cephalosporins and aztreonam regardless of the susceptibility testing results. Imipenem and piperacillin-tazobactam are the drugs of choice for successful control of such ESBL producers. Staphylococcus aureus isolates in our study were found to be uniformly susceptible to vancomycin, but were often resistant to most other antibiotics except tetracycline and erythromycin. Moreover 65.5% of them were MRSA. This is very high compared to various other studies on diabetic foot infections which have reported only 10 – 44% MRSA [5,6,7,8]. Most of the Enterococcus spp. were susceptible only to vancomycin, though they showed varying susceptibility to other antibiotics. Similarly, in another study all enterococcal isolates were noted to be uniformly susceptible to vancomycin and linezolid [5]. Hence, vancomycin can be considered as an important drug in the empirical regimen for treatment of diabetic foot infections especially in settings with high resistance to other antibiotics. In the present study 52% isolates were MDR pathogens. Earlier studies on diabetic foot infections reported 20 - 40% of the isolates to be multi-drug-resistant [7]. Most of the patients attending our tertiary care hospital have already been partially treated at various other centres and therefore exposed to several antibiotics. In addition, the widespread use of broad-spectrum antibiotics, could have contributed to the high prevalence of MDR pathogens. Similarly, in a recent study from another tertiary care hospital in India, 72% of the patients with diabetic ulcers were found to be infected with MDR organisms [5]. The main limitation of this study is the failure to detect the anaerobic bacteria. Moreover, the risk factors for the occurrence of MDR pathogens and the production of AmpC β-lactamases and metallo-β- lactamases have not been studied. A combination regimen consisting of amikacin, piperacillin-tazobactam or imipenem and vancomycin seems to be the most prudent empirical treatment of diabetic foot infection. This empirical therapy can be later modified appropriately based on the antibiogram of the isolates from the individual patients.
References |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||